Long description of the convert.pl script
1. The models
Let us start with the models. The SAM3 model is: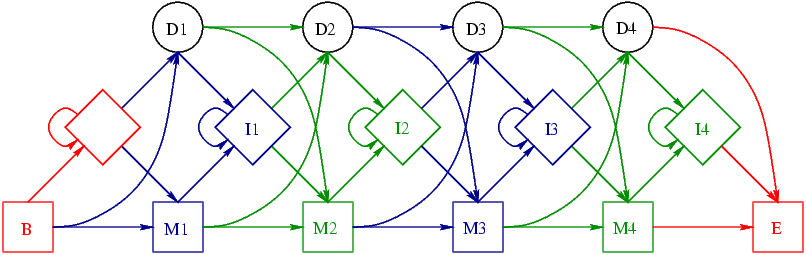
whereas the HMMER2 "Plan7" model is:
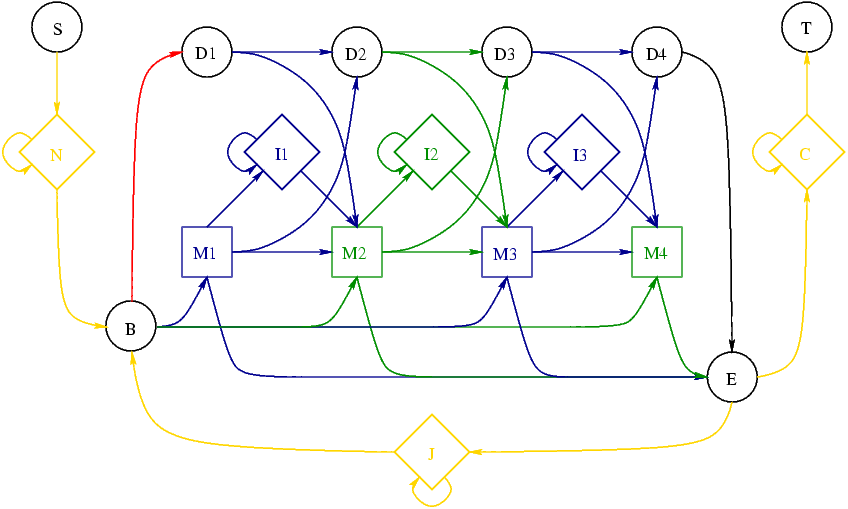
In both diagrams the circles stand for deletes, diamonds for insertions and squares for matches. The colouring shows how information is stored in the two file formats and will be discussed later. At this stage it is sufficient to note the four key differences:
- The HMMER2 model consists of a core part roughly similar to the SAM model, plus a number of additional nodes
- There are two extra insert nodes in the SAM model as compared to the HMMER core
- There are extra D->I and I->D transitions in the SAM model
- There are extra B->Mx and Mx->E transitions in the HMMER model
The first difference results in the limitations of this script: the search method (local/local etc.) is contained in the additional nodes of the HMMER model and stored in the save file, whereas in the SAM package it is specified at search-time. The authors of this script were only interested in the specific case of a local/local search and so did not include all the possibilities in the script; however, with the aid of this documentation other possibilities should not be too difficult to add.
The second and third differences result in a loss of information when converting from SAM to HMMER. (No information is lost when converting the other way.) For a sensible model of 100 or more match nodes a loss of 2 insert nodes at the beginning and end of the model should not have much effect, but the same cannot be said -- a priori -- of the loss of the insert-delete transitions. After setting these to zero in a number of SAM models and renormalising, however, we have observed no change in performance and the delete-insert transitions in the SAM model therefore appear entirely redundant.
The fourth difference does not actually result in a loss of
information when converting between SAM and HMMER, in the sense that
these transition probabilities appear to be given by a simple formula
discussed later. The possible redundancy of
these transitions was not investigated by the authors.
2. The file formats
Now to the save files themselves. The structure of a typical SAM ascii file created by thew0.5 or fw0.7 scripts
and hmmconvert is:
% : % [Some comments] % : MODEL [some details] alphabet protein FREQAVE 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.063538 0.022206 0.048871 0.060979 0.038441 0.059832 0.023936 0.060595 0.055899 0.086760 0.020237 0.045507 0.049257 0.039385 0.048255 0.071250 0.066954 0.081761 0.017985 0.038349 0.063538 0.022206 0.048871 0.060979 0.038441 0.059832 0.023936 0.060595 0.055899 0.086760 0.020237 0.045507 0.049257 0.039385 0.048255 0.071250 0.066954 0.081761 0.017985 0.038349 Begin 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.852573 0.976545 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.077669 0.016881 0.057170 0.065752 0.039223 0.062365 0.024115 0.060124 0.064073 0.082194 0.024210 0.047157 0.040425 0.041828 0.049012 0.068753 0.060345 0.072420 0.011511 0.034775 1 0.000000 0.026330 0.006903 0.000000 0.121097 0.016552 0.006787 0.002413 0.633785 0.078470 0.014492 0.040764 0.068528 0.043432 0.050361 0.044873 0.059960 0.074867 0.109772 0.028121 0.047173 0.041645 0.052252 0.020926 0.073702 0.026842 0.069371 0.010783 0.043666 0.077850 0.016640 0.057325 0.065451 0.039844 0.062177 0.024314 0.060479 0.064418 0.082109 0.024547 0.047302 0.040276 0.041960 0.048771 0.067948 0.059895 0.072042 0.011740 0.034912 2 : 3 : 4 : End 0.000000 0.000000 0.000000 0.024238 0.667485 0.022067 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 ENDMODEL
whilst a typical HMMER2.0 file created via hmmbuild -f looks thus:
HMMER2.0
NAME igs.pfam
LENG 45
ALPH Amino
:
[some non-mandatory tags]
:
XT -8455 -4 -1000 -1000 -8455 -4 -8455 -4
NULT -4 -8455
NULE 595 -1558 85 338 -294 453 -1158 197 249 902 ...
:
[some non-mandatory tags]
:
HMM A C D E F G H I K L ...
m->m m->i m->d i->m i->i d->m d->d b->m m->e
-13 * -6755
1 -2902 -5397 -782 -65 -5718 3089 142 -5469 -1006 -3213 ...
- -149 -500 233 43 -381 399 106 -626 210 -466 ...
- -17 -11924 -12966 -894 -1115 -701 -1378 -1013 -6459
2 -397 -825 529 1685 -5716 912 1172 -2586 -131 -5411 ...
- -149 -500 233 43 -381 399 106 -626 210 -466 ...
- -17 -11925 -12967 -894 -1115 -701 -1378 -6473 -6443
:
45 1367 -4042 -6533 -5898 -421 -418 -298 -2001 -5499 -408 ...
- * * * * * * * * * * ...
- * * * * * * * -6473 0
//
The files have a similar structure -- both begin with headers followed
by data for each position in the model and end with a closing tag,
ENDMODEL for SAM and // for HMMER.
The numbers in the SAM file are the actual transition/emission probabilities themselves, whereas HMMER chooses to store its integral "scores". The following formulae convert between the scores and the actual probabilities:
score = (int) floor [ 0.5 + 1000.0*log(prob/nullProb)/log(2.0) ]
prob = nullProb * 2 ^ (score/1000.0)
nullProb is the so-called null probability. To convert between
scores and probabilities, the null probability for the particular data
item must be known. (Since the logarithm of 0 is not defined, zero probabilities
are stored as score "*".)
2.1 The headers
The SAM header is rather simple, especially as theFREQAVE part is
not mandatory and according to SAM authors not used at present. The HMMER header,
on the other hand, is quite involved. It begins with the HMMER2.0 tag,
followed by a series of lines with the following structure:
[tag (5 characters)][space][data]
HMM tag. The tags must be five
characters long (shorter ones are padded with spaces at the end), so
that the data start on the 7th character of each line. The meanings
are as follows:
| Tag | Description | Null probability |
LENG |
Number of positions in the model | - |
XT |
Scores for transition probabilities between the additional (non-core) nodes of
the model (in yellow in the diagram), in the following order:
|
1.0 |
NULT |
Scores for transition probabilities of the null model (not important here) |
1.0 |
NULE |
Scores for emission probabilities of the null model in alphabetic order of the 1-letter amino-acid codes |
1/20 |
2.2 The data
The syntax of a SAM data block is:where the first 9 numbers are the state transition probabilities. The HMMER block is similar,[position number] [D->D] [M->D] [I->D] [D->M] [M->M] [I->M] [D->I] [M->I] [I->I] [20 match emission probabilities] [20 insert emission probabilities]
[position number] [20 match emission scores]
- [20 insert emission scores]
- [M->M] [M->I] [M->D] [I->M] [I->I] [D->M] [D->D] [B->M] [M->E]
nullProb=1/20.) The
columns are 7 characters wide, with the exception of the first column,
which is 6 characters wide, as in the header.
If there are M positions (data blocks) in a HMMER file,
there will be M+2 positions in the SAM file: Begin,
1, ..., M, End. The data stored in the Begin and
End blocks are shown in red in the SAM model, with the emission probabilities for the
B and E nodes being 0. Data in
consecutive blocks for both file formats are then shown in blue for
positions 1,3 and green for positions 2,4.
(There is a simple mnemonic for remembering which transition
probability is stored where: SAM stores the transition probabilities
to a particular node, HMMER stores the transition scores
from a particular node.) In the SAM file, the non-existent
transitions from M4 are stored as "*", whilst the
D4->E transition isn't stored anywhere and the authors
believe that it is implicitly set to 1.
As a final peculiarity, the B->D1 transition in SAM is stored on a
single line immediately preceding the first data block. With a null-probability of
1.0, this line has the following structure:
[ score(1-P(B->D1)) ] * [ score(P(B->D1)) ]
B->D1 probability, the
current HMMER parser demands the above syntax.
3. The program
Most of the code should be rather self-explanatory after reading the above explanations. Three notes, though: firstly, the main code at the end of the script simply decides what to do based on the extension of the filename passed to it, and secondly, the internal format of the script is essentially the SAM ascii format:where$states[$position] ->[0...8] $matches[$position]->[0...19] $inserts[$position]->[0...19]
@states holds the transition probabilities (from 0
to 8: D->D, M->D, I->D, D->M, M->M, I->M, D->I, M->I and I->I) and
@matches, @inserts the emission probabilities (0=A etc.).
$M is the number of match nodes in the model excluding the
Begin and End ones. $position=0
is the Begin position, $position=$M+1 is the
End position.
(The authors now realise that this is in fact stupid: normalising
transition probabilities say from a match node requires summing over
different positions in @states which is not particularly
elegant.)
And finally, the formulae for B->Mx and Mx->E transitions in HMMER. There seem to be some disagreements between the HMMER documentation and the actual state of affairs, so some experimentation was necessary. Check the code for actual details.